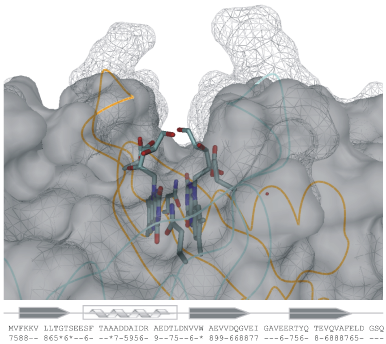
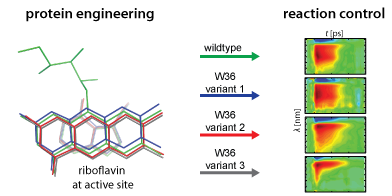
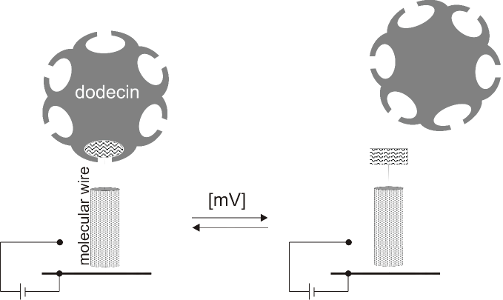Engineering structural properties of dodecin
Flavoproteins are proteins that use flavins as cofactors. The broad utilization of flavins in nature is based on the versatile reactivity of the flavins and on the proteins’ abilities to restrict this reactivity to defined reaction pathways. Protein ligand interactions mainly involve electrostatic, hydrophobic and steric interactions, but also covalent interactions. How do these interactions influence the reactivity of the bound flavin? Can these interactions be modified so that flavoprotein functions can be manipulated or engineered?
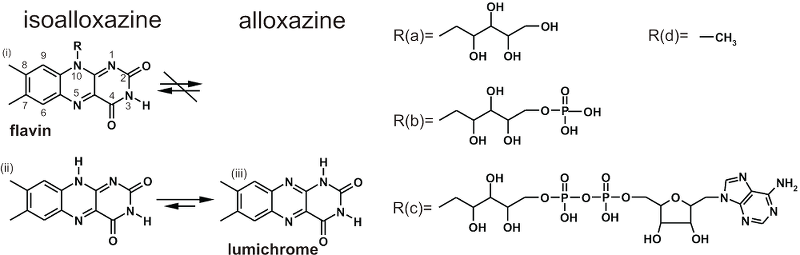
Figure: The chemistry of flavins (i) Flavins differ in the aliphatic moiety which are attached to the nitrogen N10 of a isoalloxazine skeleton (R(a)-R(d)). (ii) Lumichrome is structurally similar to flavins, but the absence of a residue R allows the 1,3-hydrogen shift to the more stable alloxazine tautomeric form with changed electronic properties.